News
Want to make a virus? It’s easy: combine one molecule of genomic nucleic acid, either DNA or RNA, and a handful of proteins, shake, and in a fraction of a second you’ll have a fully-formed virus.
While that may sound like the worst infomercial ever, in many cases making a virus really is that simple. Viruses such as influenza spread so effectively, and as a result can be so deadly to their hosts, because of their ability to spontaneously self-assemble in large numbers.
If researchers can understand how viruses assemble, they may be able to design drugs that prevent viruses from forming in the first place. Unfortunately, how exactly viruses self-assemble has long remained a mystery because it happens very quickly and at very small length-scales.
Now, there is a system to track nanometer-sized viruses at sub-millisecond time scales. The method, developed by researchers at the Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS), is the first step towards tracking individual proteins and genomic molecules at high speeds as they assemble to create a virus.
The research was led by Vinothan Manoharan, the Wagner Family Professor of Chemical Engineering and Professor of Physics, and was published recently in ACS Nano. Manoharan’s group worked in collaboration with researchers at Leiden University, MIT, the Leibniz Institute of Photonic Technology, the University of Jena, and Heraeus Quarzglas, a manufacturer of fiber optics.
“Our goal is to understand how viruses manage to assemble spontaneously, so quickly and so robustly,” said Yoav Lahini, research associate, former Pappalardo Fellow at MIT, and co-first author of the study.
Identifying critical intermediate stages in the assembly process could help researchers understand how to interfere with this process, Lahini said. Shedding light on the physics of self-assembly could also help engineers design better synthetic nanomaterials that can spontaneously piece themselves together.
There are two main challenges to tracking virus assembly: speed and size. While fluorescent microscopy can detect single proteins, the fluorescent chemical compound that emits photons does so at a rate too slow to capture the assembly process. It’s like trying to observe the mechanics of a hummingbird’s flapping wing with stop-motion camera; it captures pieces of the process but the crucial frames are missing.
Very small particles, like capsid proteins, can be observed by how they scatter light. This technique, known as elastic scattering, emits an unlimited number of photons at a time, solving the problem of speed. However, the photons also interact with dust particles, reflected light, and imperfections in the optical path, all of which obscure the small particles being tracked.
To solve these problems, the team decided to leverage the outstanding quality of optical fibers, perfected over years of research in the telecommunication industry. They designed a new optical fiber with a nano-scale channel, smaller than the wavelength of light, running along the inside of its silica core. This channel is filled with liquid containing nanoparticles, so that when light is guided through the fiber’s core, it scatters off the nanoparticles in the channel and is collected by a microscope above the fiber.
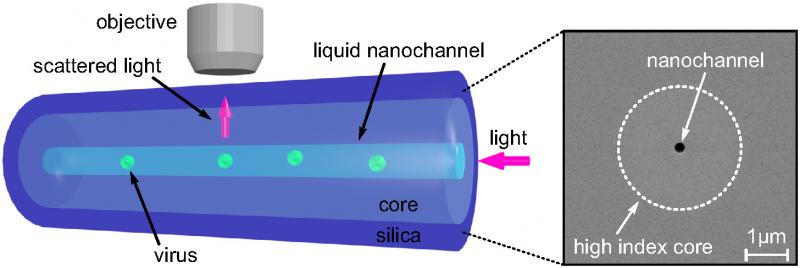
Optical fiber with a nano-scale channel
The researchers observed the motion of viruses measuring 26 nanometers in diameter at a rate of thousands of measurements per second.
“These are the smallest viruses to be tracked on sub-millisecond time scales, which are comparable to the time scales for self-assembly.” said Rees Garmann, post-doctoral fellow in the Manoharan lab and co-author of the research.
The next step is to track not just single viruses but single viral proteins, which scatter 100 to 1,000 times less light than a single virus.
“This research is a step forward in observing and measuring the self-assembly of viruses,” said Manoharan. “Viral infection involves many complex molecular and cellular pathways, but self-assembly is a process that is found in many different viruses. This simple technology, which is cheap, easy and scalable, could provide a new, cost effective way to study and diagnose viruses. From the point of view of fundamental physics, understanding the self-assembly of a naturally evolved system would be a major milestone in the study of complex systems.”
Topics: Health / Medicine, Applied Physics
Cutting-edge science delivered direct to your inbox.
Join the Harvard SEAS mailing list.
Scientist Profiles
Vinothan N. Manoharan
Wagner Family Professor of Chemical Engineering and Professor of Physics
Press Contact
Leah Burrows | 617-496-1351 | lburrows@seas.harvard.edu




