News
Plastic water bottles that end up as litter contribute many of the 5 trillion tiny plastic pieces that are polluting the world's oceans. (Photo courtesy of the National Oceanic and Atmospheric Administration.)
More than five trillion tiny pieces of plastic are floating on the surface of the world’s oceans, threatening marine life and posing health hazards to humans. Bacteria could help researchers detect and clean up these plastic fragments, according to research conducted by a group of Harvard undergraduates.
The students, members of the International Genetically Engineered Machine (iGEM) team at the John A. Paulson School of Engineering and Applied Sciences (SEAS), used synthetic biology to tackle the problem of marine micro-plastic pollution. Their solution, Plastiback, won a gold medal at the annual iGEM competition, which included more than 300 student teams from dozens of nations. It was also nominated for best environmental project in the undergraduate category.

The Harvard team at the International Genetically Engineered Machine (iGEM) Competition championship in Boston. (Photo courtesy of the Harvard iGEM team.)
“Ocean researchers don’t currently have a very good way of quantifying or detecting where these tiny pieces of plastic may be in the ocean,” said team member Lorena Lyon, A.B. ’18, a human development and regenerative biology concentrator. “Current methods typically involve trawling the ocean, which means researchers go out with a net and physically count the pieces of plastic they pick up. That is not very efficient.”
Plastiback is a floating ocean bioreactor containing genetically modified E. coli bacteria, which is used to identify and degrade a type of micro-plastic called polyethylene terephthalate (PET). PET is commonly used in many products that end up as ocean pollution, from disposable water bottles to commercial fishing equipment. The students genetically engineered these E. coli to secrete an enzyme that breaks down PET into terephthalic acid and ethylene glycol. A fluorescent protein glows green when the PET-degrading enzyme is expressed, so researchers who use the device can easily see whether the system is working correctly.
“Degrading plastic in the ocean leads to less plastic, which is a great thing up to that point, but the byproducts of plastic are harmful, so we added another step to our project to extend that idea of environmental remediation,” said Kevin Shani, S.B. ’18, a bioengineering concentrator.
During the next step, the terephthalic acid produced during degradation flows into a microbial fuel cell located in another chamber of the floating bioreactor. A second type of bacteria, D. tsuruhatensis sp. nov., metabolizes the acid, producing electrons that are used to generate a small amount of voltage. Researchers could measure the voltage to quantify how much PET was broken down by Plastiback.
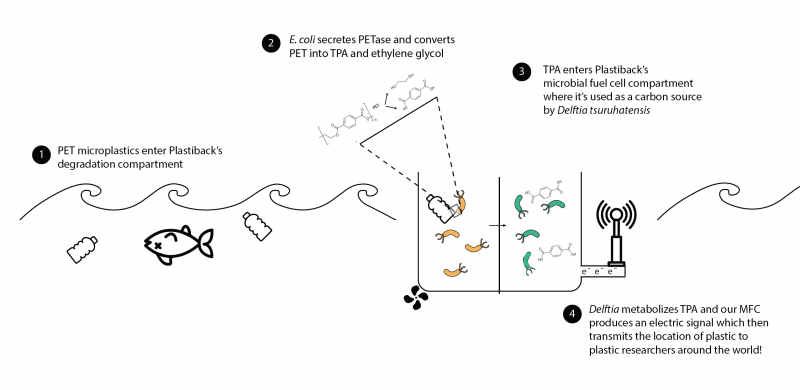
This diagram shows how the Plastiback process works. (Image courtesy of the Harvard iGEM team.)
Not only would Plastiback help degrade harmful plastic in the oceans, but its detection system could also enable researchers to create an ocean map pinpointing “hot spots” where large concentrations of micro-plastic exist, Lyon explained.
“These micro-plastics are so dangerous because they can absorb toxins, like DDT, and then be consumed by fish,” said Yasmeen Fakhro, A.B. ’18, a chemistry and physics concentrator. “If those fish are eaten by humans, that poses a huge risk to public health.”
A recent study published in Nature found that more than 25 percent of fish for sale in public markets contained micro-plastic. Identifying large collections of micro-plastic could help fishermen and food regulators recognize public health dangers that may be present in each catch.
As part of the iGEM competition, the students submitted the E. coli genetic constructs they developed to the Registry of Standard Biological Parts, making it possible for researchers around the world to access the data and apply it to their own research.
While certain parts of the project had a steep learning curve, such as retrofitting a microbial fuel cell, working as a team to tackle an important environmental issue was rewarding, said Daniel Um, A.B. ’19, an integrative biology concentrator pursuing a secondary in computer science.
“This field has been growing exponentially over the past decade,” he said. “It is exciting to be part of this wave of synthetic biology research. I’m proud of what we were able to accomplish.”
The team, which began working on their project over the summer, also included students Gabriela Berner, Will Cho, Rebekah Chun, and Mofeyifoluwa Edun. They were mentored by Neel Joshi, Associate Professor of Chemical and Biological Engineering, and graduate students Pichet Praveschotinunt, David Lips, Kevin Hof, and Anita Chandrahas.
"The team deserves a lot of credit for designing, planning, executing, and presenting a project that they conceived primarily on their own, with some help from the mentoring team,” said Joshi. “I hope that the success of this year's team can continue to attract more undergraduates to participate in the team and in the student group started by last year's team."

Get involved with iGEM
Harvard undergraduates who are interested in getting involved with iGEM on campus can contact crimsonigem@gmail.com. In addition to participating on the summer iGEM research team, the Harvard College iGEM club offers opportunities to visit biotech companies, hear world-class speakers, and pursue student-directed research. This Wintersession, the club is hosting a BioDesign Bootcamp in which students can discover synthetic biology (no prior experience necessary). Apply now at http://bootcamp.harvardigem.com.
Topics: Bioengineering
Cutting-edge science delivered direct to your inbox.
Join the Harvard SEAS mailing list.
Press Contact
Adam Zewe | 617-496-5878 | azewe@seas.harvard.edu



